What is immunohistochemistry (IHC) and what is it used for?
Immunohistochemistry is a process for detecting specific proteins in sections of biological tissue using antibodies. It can be used for a variety of applications in the biomedical sciences, such as characterizing diseases, assessing disease progression, and studying molecular processes at different developmental stages.
What are the typical steps in an IHC protocol?
The steps of conventional IHC broadly consist of: fixing tissue, sectioning the tissue, staining for targets of interest, and imaging sections under a microscope. The details of these steps depend on the method of antibody detection: the two most commonly-used methods are chromogenic and fluorescent IHC (AKA immunofluorescence). Chromogenic IHC uses enzymatic reactions to stain tissue, while fluorescent IHC relies on fluorophore-conjugated antibodies.
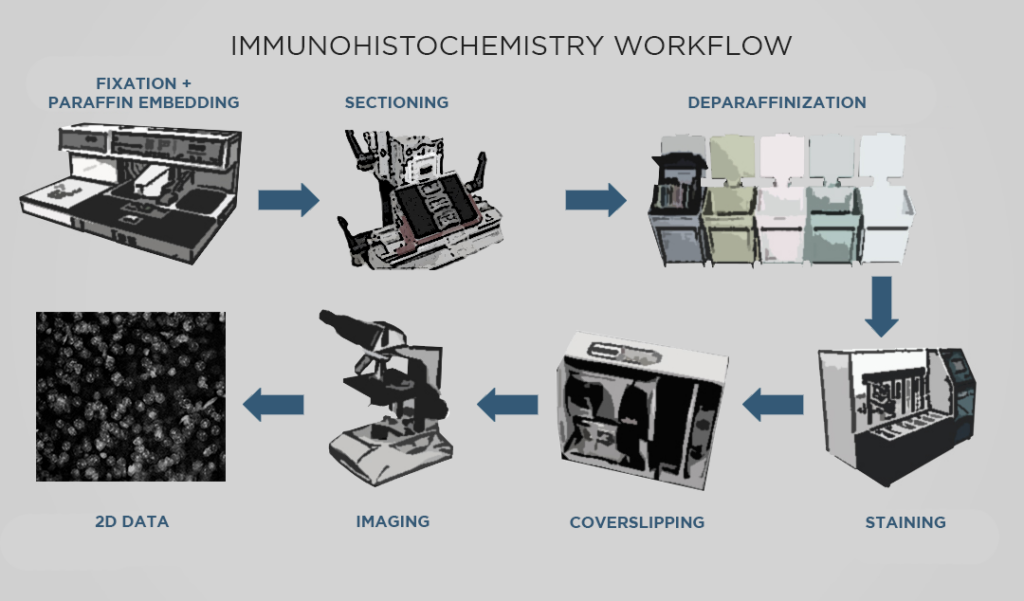
For both processes, the tissue typically goes through formalin fixation. Fixing tissue with formalin (AKA formaldehyde) creates crosslinks that stabilize important features like proteins, nuclei, and so on, so that those features are relatively preserved in future steps. Then, if thin slices are required, the fixed sample is infiltrated with paraffin which produces a formalin-fixed, paraffin-embedded (FFPE) sample that is hardened and suitable for sectioning.

Mouse brains sectioned into 500 µm (left) and 200 µm slices (right) with Megatome.
Next, the sample is cut into slices. For chromogenic IHC, the sample must be cut into very thin slices with a microtome, allowing light to effectively pass through the sample with a brightfield microscope. For immunofluorescence, the tissue can be cut into thicker sections with a vibrating microtome. Sectioning also reduces the diffusion distance so that antibody probes can effectively reach their targets. Paraffin-embedded samples must go through a process called deparaffinization before they can proceed to the next step.
Then, IHC staining involves incubating the sections with one or more probes depending on the desired targets. Staining allows for selective visualization of proteins in the sample. In chromogenic IHC, detection is accomplished by an enzymatic reaction that converts a substrate such as diaminobenzidine (DAB) into a colored stain. In immunofluorescence, the fluorophores conjugated to antibodies allow visualization under a fluorescence microscope.
Finally, the sample sections are mounted on slides and imaged with a microscope to generate visual data showing protein expression, which can then be analyzed qualitatively or quantitatively. Samples processed with chromogenic IHC are typically imaged with a brightfield microscope, as the chemically-induced stains are visible under transmitted light. Samples processed with fluorescent IHC must be imaged with a fluorescence microscope, such as a confocal microscope.
What are the challenges of using IHC methods to process samples and generate data?
While immunohistochemistry has been used in biomedical research for decades, the methods involved have many shortcomings which newer techniques have addressed. First, conventional IHC inherently generates two-dimensional data because of the need to section samples into thin slices. This significantly impacts data quality, as biological processes are spatially complex and cannot be fully or accurately captured in 2D. The figure below shows how different phenomena appear in 3D versus 2D, highlighting how important spatial context is to collecting and interpreting data.
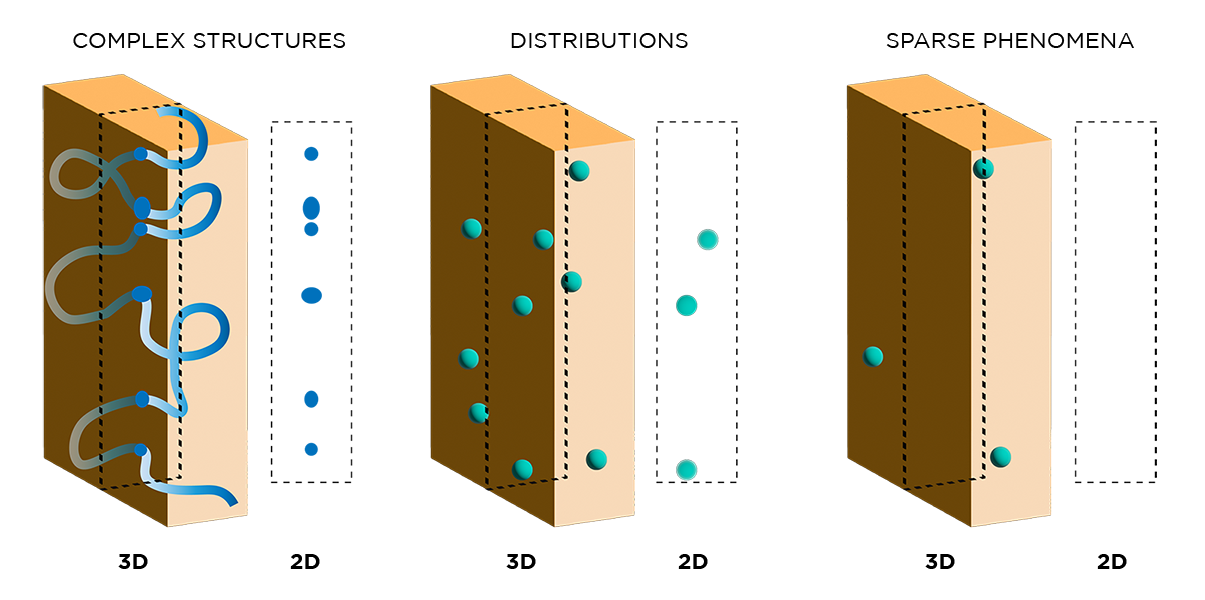
"...Biological processes are spatially complex and cannot be fully or accurately captured in 2D."
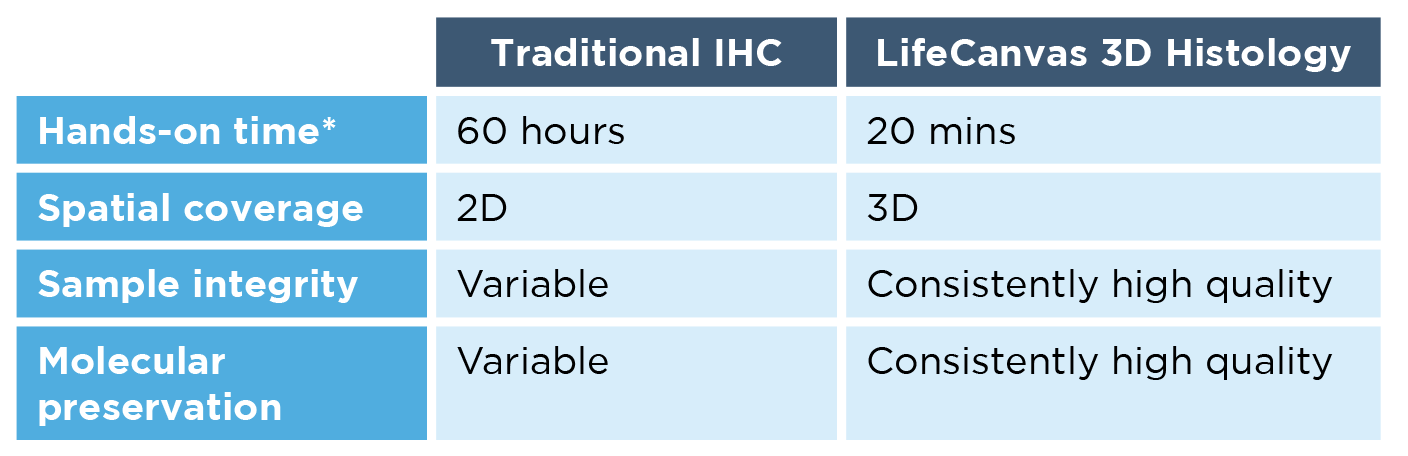
Second, preparing and imaging numerous thin tissue sections is tedious and time-consuming – sample processing alone can take up to 5 hours of hands-on time per sample. Additionally, as experimental steps and number of sections increase, so does the potential for damaging samples.
Third, 2D sectioning and imaging leads to downstream sampling issues that can also affect data quality. Even if every section safely makes it through processing, it could take a prohibitive amount of time to image all of the samples. Typically, a subset of high-quality sections are imaged and used for subsequent analysis. Computer software can be used to create 3D reconstructions from stacks of this 2D data, however these reconstructions will vary in accuracy depending on the number and quality of sections chosen for imaging.
What is 3D histology and how does it differ from IHC?
3D histology refers to the process of preparing intact tissues for imaging – instead of slicing samples, experiments are performed on whole organs. Samples prepared in this way are then imaged intact using light sheet microscopy, generating spatially unbiased and informative 3D data. This approach requires a different suite of tools and techniques, as traditional IHC methods are not compatible with thick tissues. At LifeCanvas, we have spent years developing and optimizing our end-to-end 3D histology pipeline to facilitate this process for researchers.
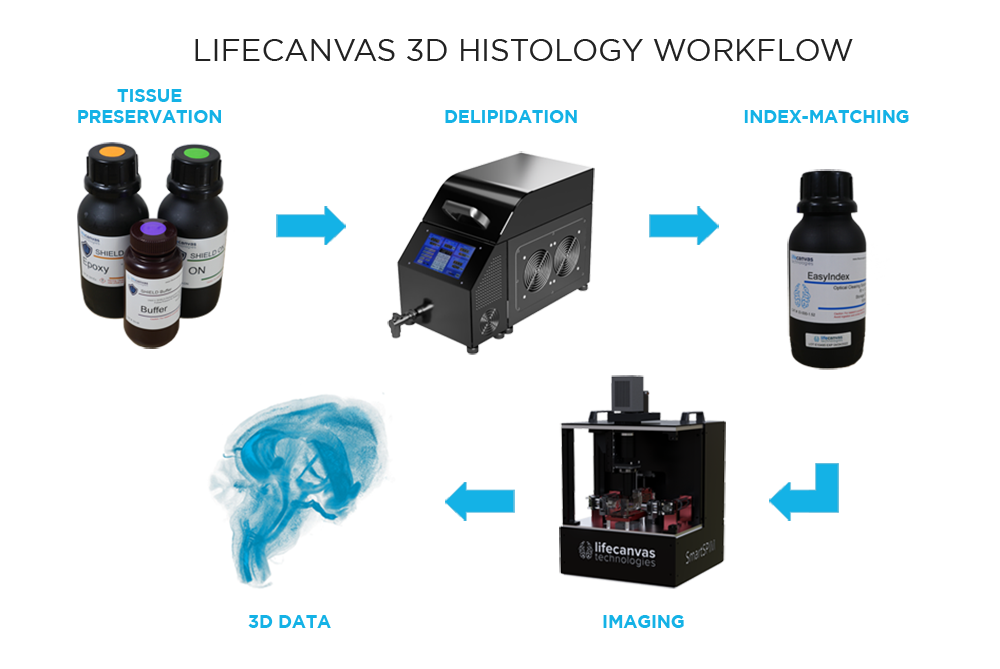
What are the steps in our 3D histology pipeline?
Our workflow begins with SHIELD tissue preservation, a Nature Biotechnology-published technique that further stabilizes the key molecular features of whole, formaldehyde-fixed tissues. This epoxidation-based method protects tissue architecture, antigenicity, molecular probes, structural features, and endogenous proteins during downstream processing.
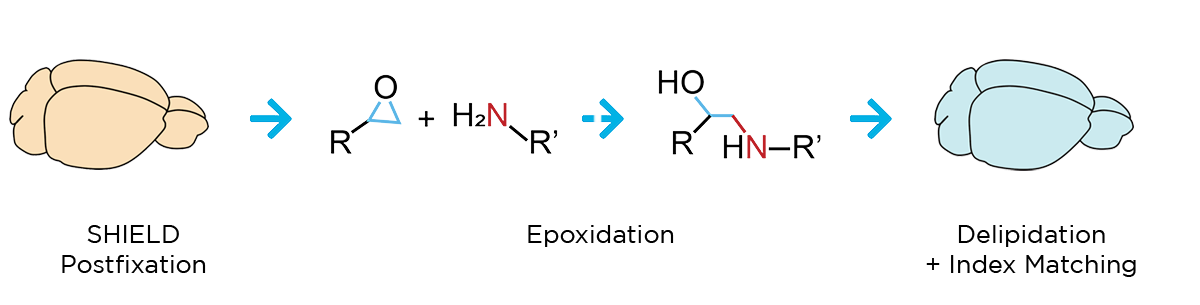
Next, a process called delipidation is necessary to allow optimal light penetration into the thick tissue sample during imaging. During delipidation, our Clear+ buffers gently remove light-scattering lipids from the tissue without impacting sample morphology or fluorescent proteins. This can be done in just one day using our patented active clearing technique, or in about a week with passive methods for mouse organ-sized samples.
Because of our unique combination of SHIELD preservation and Clear+ delipidation, endogenous fluorescent proteins are protected and do not need to be labeled with antibodies. Additional immune targets can be labeled with our patented active labeling technique, which achieves uniform, robust probe penetration throughout the tissue in just 1-2 days. Our SmartBatch+ device combines active tissue clearing and labeling functionality to further streamline the 3D histology workflow. Accommodating up to 12 whole mouse brains or 4 whole rat brains, SmartBatch+ enables high-throughput clearing and labeling with just a few simple buffer changes and button clicks.
*For 12 whole mouse brains
What challenges introduced by IHC does our pipeline solve?
By processing (and subsequently imaging) whole specimens in 3D, our histology techniques overcome many of the challenges of traditional immunohistochemistry. First, 3D data generated with our techniques more accurately represent spatially complex phenomena such as continuous structures, connectivity, cell distributions and interactions, and rare events.
Second, our workflow is much faster, simpler, and more scalable than IHC. Using the SmartBatch+ system, the LifeCanvas 3D histology pipeline requires just 20 minutes of hands-on time – regardless of whether you’re processing 1 or 12 whole mouse brains. This not only speeds up the workflow by orders of magnitude, but ensures experimental consistency across samples.
"3D data generated with our techniques more accurately represent spatially complex phenomena such as continuous structures, connectivity, cell distributions and interactions, and rare events."
Third, working with whole organs bypasses the sampling issues inherent to IHC and section-based histology. Light sheet microscopy captures high-resolution volumetric data from the entire tissue, which can then be analyzed with our 3D image data analysis tools. This provides a detailed, unbiased look into the biological processes of interest.
To learn more about transitioning from IHC to 3D histology, check out our white paper or directly get in touch with one of our scientists.

