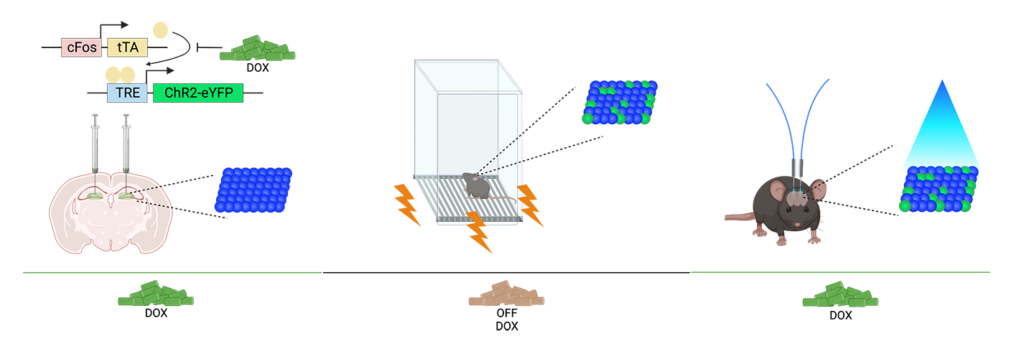
Updated February 27, 2023: Kaitlyn’s preprint “Hippocampal engrams generate flexible behavioral responses and brain-wide network states” is now available on bioRxiv.
Kaitlyn Dorst is a Ph.D. candidate in the Ramirez lab at Boston University.
LifeCanvas (LC): What has been the focus of your dissertation work?

I wanted to hone in on how this engram produces different behavioral phenotypes when I modify external variables, such as the animal’s environment size. Even though the mice all experience the same fear engram reactivation, they behave differently in different environment sizes. Another element of the project is using a brain-wide approach to see what is happening beyond the hippocampus during engram manipulation, and what brain regions may be interacting to produce a phenotype.
LC: Do mice respond differently to fear engram reactivation versus naturally recalling the fear memory?
KD: There is variability in their behavior during the initial fear conditioning. Some rodents become more fearful, while others are more resilient. However, with optogenetic manipulation, they produce a more consistent freezing phenotype within the same environment size. While naturalistic memory may engage the same populations of cells as optogenetic manipulation, it may not produce as strong of an effect – we’re really stepping on the gas pedal to hijack how these memory ensembles work.
We do have network data to analyze from LifeCanvas looking at differences between mice experiencing natural recall versus optogenetically-induced recall. These mice go through fear conditioning in certain chambers, and are placed back in those chambers 24 hours later.
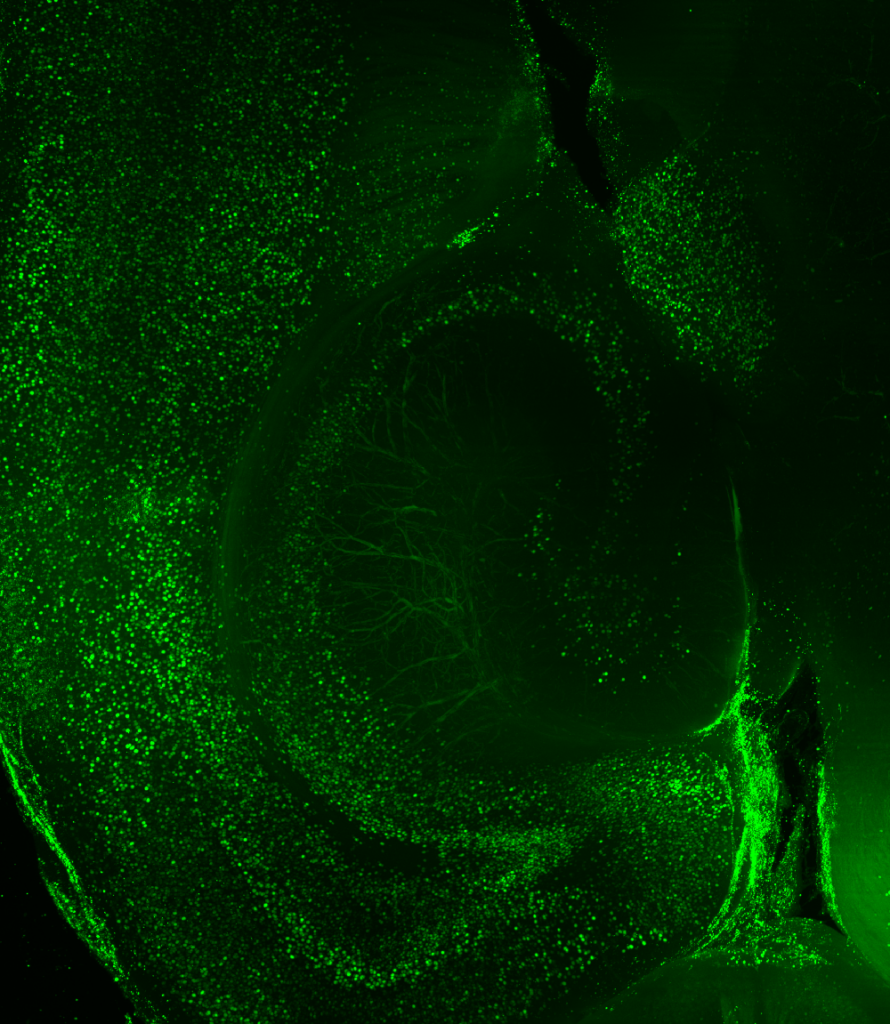
LC: How have LifeCanvas’ tools and services helped you address your research questions?
KD: Before working with LifeCanvas, my approach consisted of looking at coronal sections with a confocal microscope. But for the second part of my project, examining brain-wide interactions upon artificial manipulation of hippocampal neurons, I really needed to take a brain-wide approach. I was very excited to send whole brains to LifeCanvas for endogenous c-FOS clearing, immunolabeling, light sheet imaging, and analysis.
"Without a whole-brain approach, we may have missed entire thalamic and hypothalamic areas that appear to act as hubs."
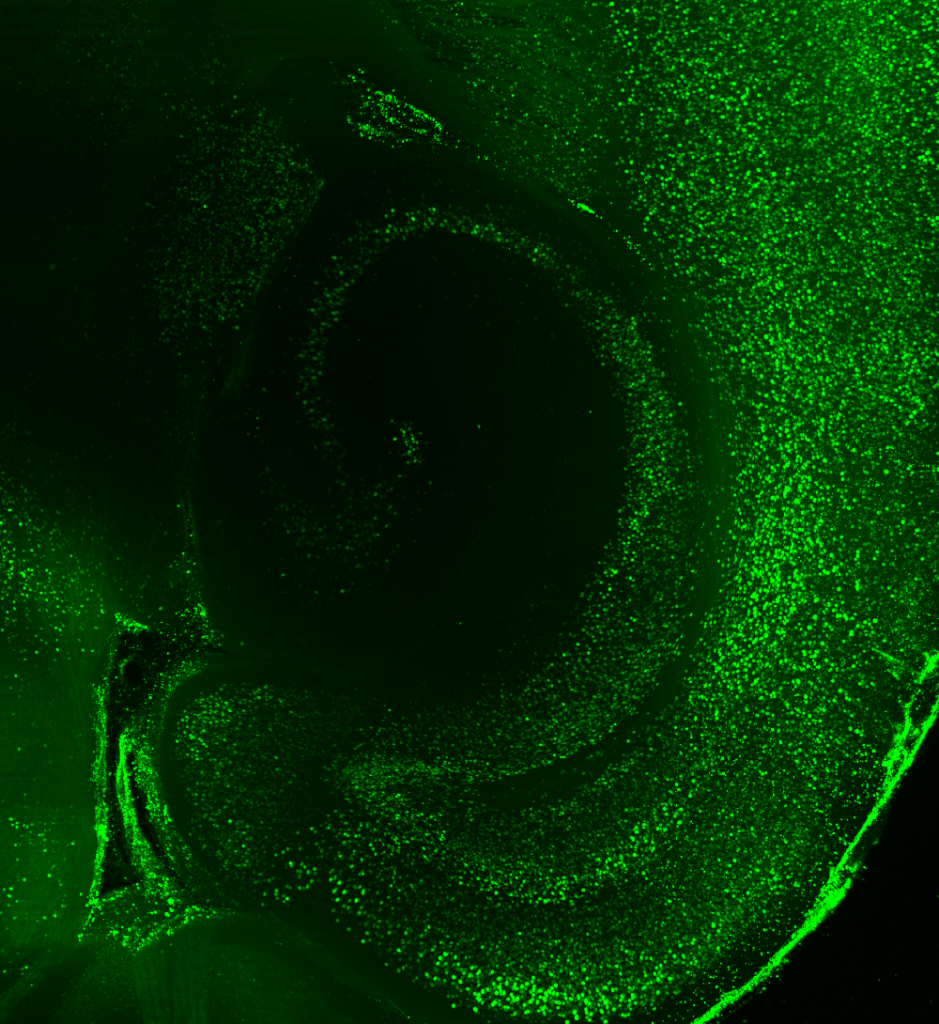
LC: How do your analytical approaches provide insight into functional connectivity?
KD: We generate correlations across 8 animals and 147 brain areas to see how c-FOS levels are consistently or differentially altered. We then plot these correlation values on graphs, showing regions of the brain as nodes, and correlations between those regions as edges. We can then examine the structure of the graph at the mesoscale: what are the trends in all the brain regions and their connections, as well as between optogenetically-modified and control animals?
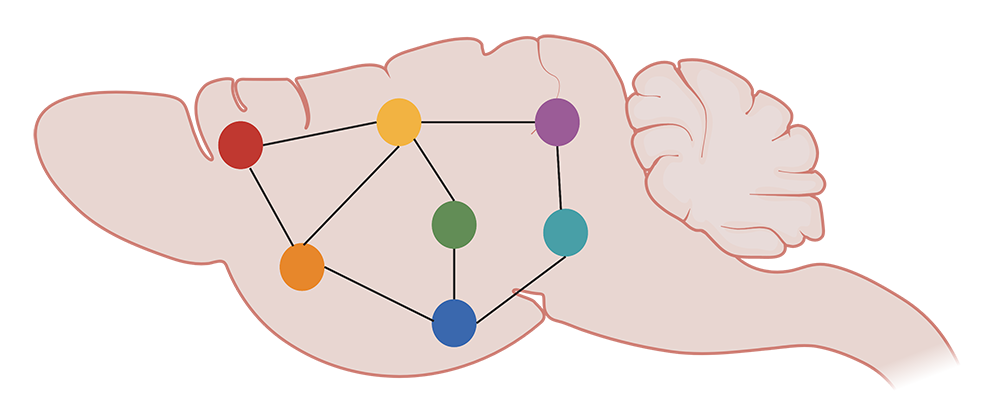
We can get more information on these trends by functionally segregating the data with a clustering algorithm, which essentially groups brain areas by correlation profile. These are regions that respond to reactivation similarly. Eventually, we also want to quantify differences in the topology of these networks, as well as zoom in on the contributions of individual brain areas. This can be done by running simulations to, for example, delete hub regions from the network and see how the edges potentially reshuffle.
LC: What are the translational implications of this research?
"... If we can find hub regions of the brain that are highly interconnected within these networks, we could potentially identify new therapeutic target areas."
KD: Literature shows that optogenetics can be used to bring a tagged memory back online in an Alzheimer’s mouse model, suggesting that this memory may not be erased, but is just somehow inaccessible. On the flip side, with a condition like PTSD, you could potentially quell an active memory.