The goal of histology has long been to interrogate complex structures in whole tissue samples. Over centuries, slicing of samples for traditional two-dimensional histology has been indispensable in understanding the structure and morphology of multiple tissue types. However, this approach is limited to yielding images in thin tissue sections, is time-consuming and is error-prone due to tissue distortion and information loss.
Current research demonstrates the significance of using state-of-the-art clearing techniques and three-dimensional volumetric imaging in examining tissue-wide connectivity and cell distribution in a standardized high-throughput manner.
A recent study “Delineation of an insula-BNST circuit engaged by struggling behavior that regulates avoidance in mice”, published in Nature Communications by Luchsinger et al. (Vanderbilt University School of Medicine, TN), describes an insular cortex to bed nucleus of the stria terminalis (BNST) neuronal circuit that controls affective behavior in mice after inducing struggling behavior. Dr. Joe Luchsinger and colleagues conducted these studies in the developmental neurobiology laboratory of Dr. Rich Simerly (Simerly Lab) at the Vanderbilt Center for Addiction Research with lightsheet imaging and analyses provided through the recently established Vanderbilt Neurovisualization Lab managed by Dr. Jose Maldonado.
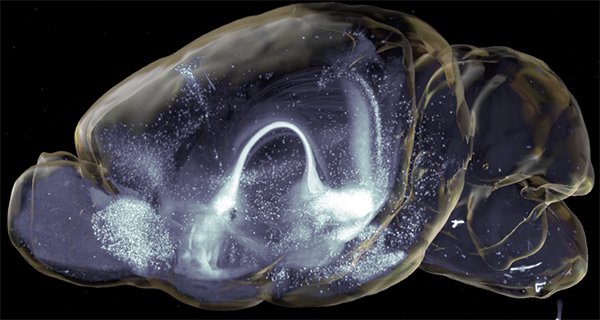
A key experiment involved retro- and anterograde viral tracing strategies to map neuronal connections in whole brains to assess the afferent network for insula→BNST neurons. For this, Dr. Luchsinger prepared the brains according to the SHIELD protocol and performed active clearing using SmartClear with four whole brains at the same time.
Importantly, through this approach Dr. Luchsinger was able to clear and image whole brains in a standardized, robust way without the need of tissue sectioning and without the loss of critical information.
Next, in order to illustrate the efficiency of assessing brain-wide circuitry in three-dimensions, Dr. Luchsinger performed quantification of cell counts in whole brain volumes and demonstrated the distribution of labeled neurons in the insula→BNST control network in a semi-autonomous manner using custom modified software, including SmartAnalytics.
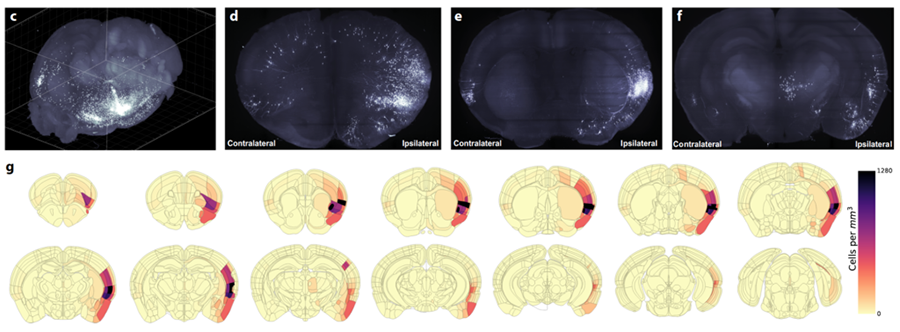
He further added: “We uncovered an unexpectedly large input from executive motor cortical regions and demonstrated that activity in these insula-projecting motor cortex neurons is time-locked to and precedes the onset of the restraint stress struggle events and insula→BNST activity.”
In another recent publication, “The melanocortin-3 receptor is a pharmacological target for the regulation of anorexia”, published in Science Translational Medicine by Dr. Patrick Sweeney (University of Michigan) & Dr. Michelle Bedenbaugh (Vanderbilt University School of Medicine) further demonstrated how whole tissue clearing and 3D imaging advanced their research through a collaborative study between the labs of Dr. Simerly (Vanderbilt) and Dr. Roger Cone (University of Michigan).
Initially, she pretreated the brains with SHIELD (protecting endogenous fluorescence) prior to active clearing with SmartClear. Importantly, this avoids an immunolabeling step, which not only saved time but also unnecessary antibody expenses. After refractive index matching with EasyIndex, brains were again imaged using SmartSPIM light sheet microscopy.
Dr. Bedenbaugh defined clusters of MC3R-positive neurons throughout the hypothalamic areas, including those that were previously established to be implicated in regulating food intake, e.g. ventromedial and dorsomedial hypothalamus.
This streamlined approach enabled fast characterization of MC3R+ cell distributions and complemented the pharmacological and genetic approaches to uncover the MC3R as a promising therapeutic target for treating anorexia-related disorders and weight loss.
Conclusion
Recent studies show the value of using whole tissue clearing and 3D imaging to investigate whole brain neuronal circuitry and cell distribution.
Our products enable:
- Fast, standardized, high-throughput results
- Whole tissue quantification without loss of information
- Whole tissue visualization that increases the chance of discovering novel findings