
LifeCanvas (LC): What did you work on prior to joining the Allen Institute?
Mike Taormina (MT): I got my Ph.D. in Physics at the University of Oregon, where I initially became interested in light sheet microscopy. There, I used it specifically for live specimen imaging in zebrafish – at the time, light sheet microscopy was mostly used for live imaging over long periods of time. It’s exciting to see this technology now being used in large organ samples and cleared tissues.
LC: Where has your research taken you at the Institute?
MT: I’m currently a scientist in the imaging department. My team supports a wide range of projects in brain science, from studying cell connectivity and morphology, to characterizing novel transgenic lines. However, at the Allen Institute, each team operates somewhat like a research core. Samples can move through different groups, from lab animal services and transgenic colony management teams all the way up to informatics and data management. As an imaging scientist, I get to work at this boundary between providing core services and being involved in more technical projects, such as those involving whole-brain light sheet imaging.
LC: How has whole-brain imaging technology evolved at the Allen Institute over the years?
"We tested many solutions ... for both clearing and imaging tissue. LifeCanvas’ tools reliably generate better results more quickly than many other techniques."
MT: The Allen Institute has been using a serial sectioning pipeline since about 2011, which has produced around 11,500 datasets (with each dataset comprising a series of images through a whole mouse brain). This pipeline has had a significant impact – for example, it produced the anatomical template for the Allen reference brain atlas, and also provided the data for our connectivity atlas.
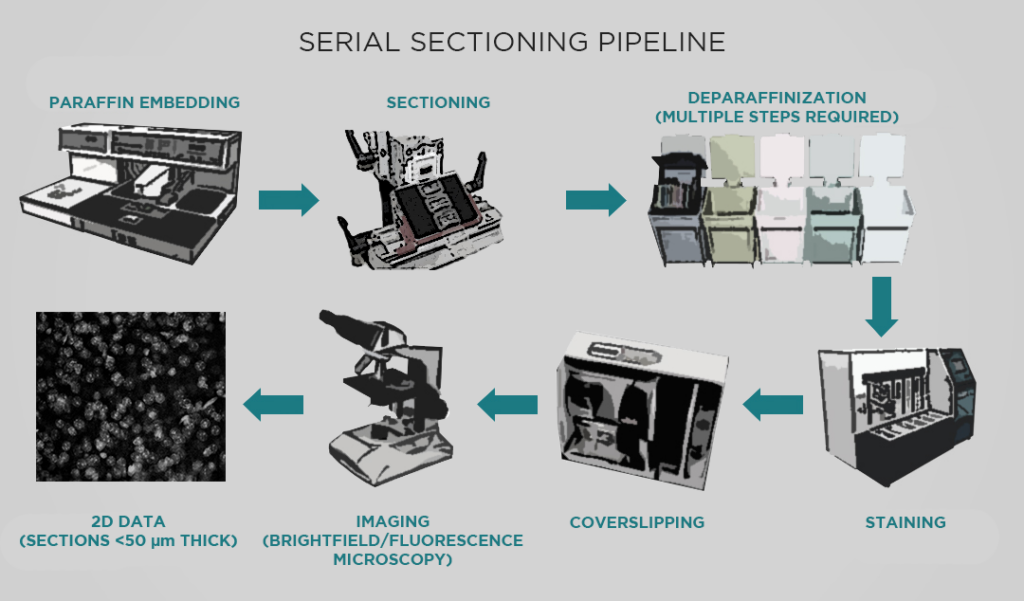
However, this pipeline has many limitations. For one, a series of sections is usually sparsely sampled. Taking 140 sections through a mouse brain produces a very large dataset, but we miss a lot of data in the 100 microns between sections. With light sheet microscopy, we can sample more or less isotropically, obtaining truly volumetric data. We’re excited to use light sheet to build atlases for more specific types of projects – for example, developing viral genetic tools to target specific transcriptomic cell types in living specimens.
LC: How have LifeCanvas tissue clearing and imaging tools helped the Institute adopt this volumetric imaging approach?
MT: We want to gradually replace this serial sectioning pipeline with one that can ideally run for at least another 10 years and 10,000 samples. We tested many solutions, both commercial and noncommercial, for both clearing and imaging tissue. LifeCanvas’ tools reliably generate better results more quickly than many other techniques.
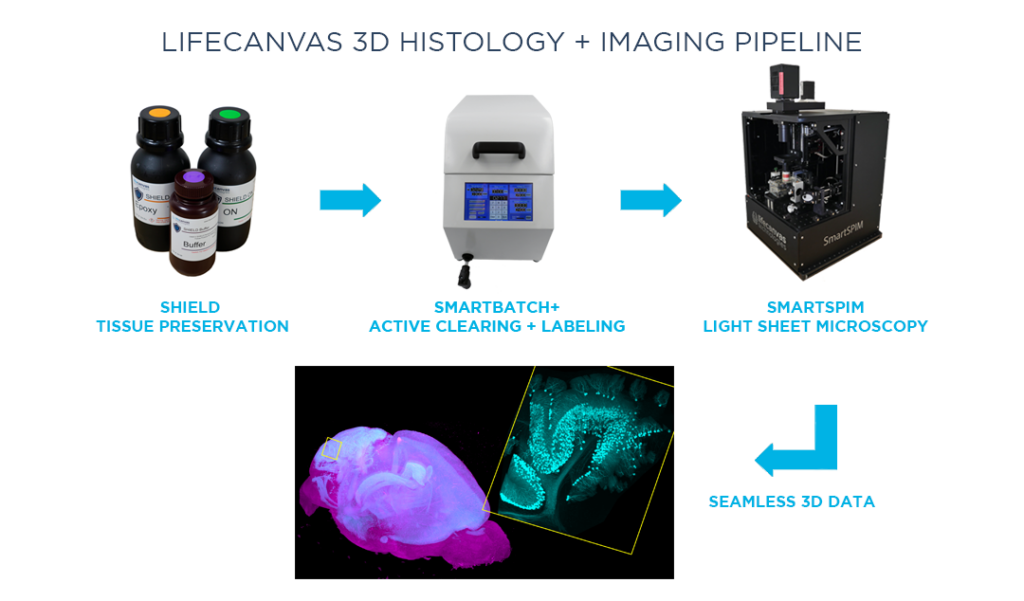
Besides consistently producing high-quality data, we also wanted a system that would be relatively straightforward to adopt in our lab. In the world of light sheet microscopy, running the instrument itself and mounting samples have always been pain points. Everything grinds to a halt after the one scientist who knows all of that particular system’s secrets leaves. In contrast, we can quickly train researchers to clear samples with SmartBatch+ and image samples with SmartSPIM, and get good data every time.
LC: How has SmartBatch+ tissue clearing helped facilitate whole-brain imaging experiments?
MT: We were seeking a tissue clearing technique that would preserve fluorescent proteins and native fluorescent signals. We work predominantly with samples from transgenic animals with endogenous fluorescent proteins, or fluorescence induced by a viral injection. Many tissue clearing methods are based on organic solvents, which kill fluorescent protein signal and thus necessitate labeling samples. LifeCanvas’ tissue clearing techniques and the SmartBatch+ device protect endogenous fluorescence, saving us a lot of time, labor, and resources.

LC: Besides the added spatial information from 3D imaging in general, what are specific advantages of SmartSPIM light sheet microscopy?
MT: Throughput was a huge priority for us. SmartSPIM is faster than competitors, sometimes by an order of magnitude. There’s a significant difference between being able to image 4 brains a day versus one brain every 2 days. Our serial sectioning pipeline comprises 6 instruments, which have the capacity to process roughly 1200 brains in a year. Even then, we still get a backlog – there’s never a shortage of people sending us samples labeled “ASAP”. One SmartSPIM can easily replace 3 of those systems.
Additionally, SmartSPIM allows us to easily separate multiple colors in one experiment, while the serial sectioning instruments are limited by 2-photon fluorescence excitation. Having many colors is incredibly useful for scaling up experiments. The SmartSPIM at the Institute for Neural Dynamics has 5 laser lines, which allows us to easily distinguish 5 different fluorophores in one cleared sample. This further increases the throughput by allowing us to not only image one brain more quickly, but to also do more experiments in one brain.
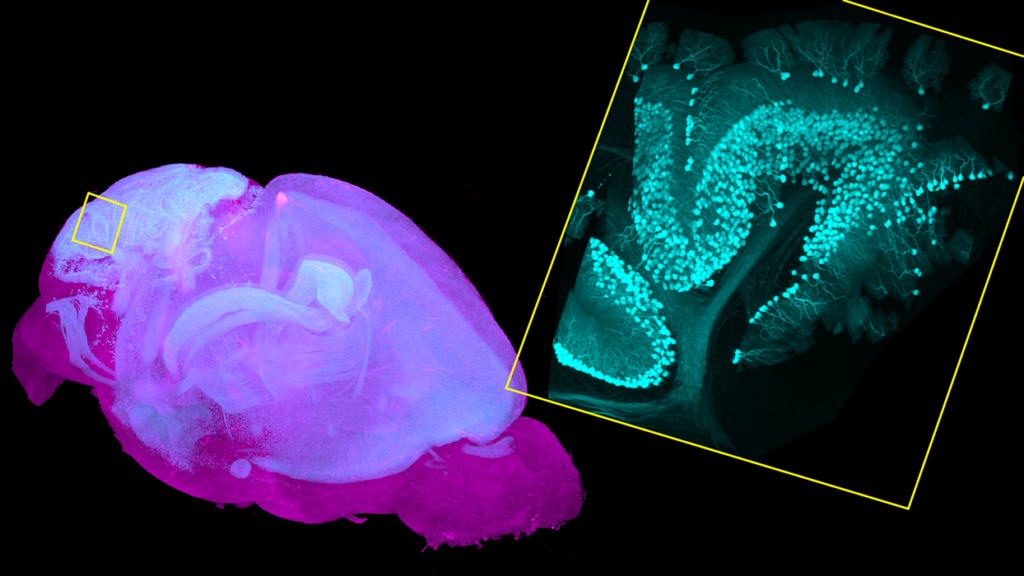
LC: Imaging whole brain samples in 3D notably produces a lot of data. How do you analyze and make sense of it?
MT: The Allen Institute has always had an open science, open data mentality and commitment. Our scientific computing team has been building a system to not only host the raw data from this new pipeline, but also provide rich, extensive metadata describing the sample and how it was processed. We want to package this information on the cloud in a way that includes both the software tools and the environment in which those tools are run, so that people can easily rerun analyses or insert their own.
LC: What applications of this whole-brain light sheet data excite you?
MT: In addition to generating better comprehensive atlases – for different developmental time points or model animals, for example – we will use whole-brain light sheet imaging to quantitatively characterize expression patterns of the transgenic or viral genetic tools we are developing. Quantitative description of the expression patterns of these tools, which are used to label specific cells in specific parts of the brain, is currently limited.
"SmartSPIM is faster than competitors, sometimes by an order of magnitude ... [and] allows us to easily distinguish 5 different fluorophores in one cleared sample."
We plan to use the SmartSPIM microscope to acquire this brain-wide data and register the datasets to our Allen Institute Mouse Common Coordinate Framework, enabling quantification of the numbers and locations of cells for each genetic tool.
This data will be integrated with other spatial or molecular information so that researchers will be able to make an educated decision about which tool to select to investigate a specific brain region or cell type. This process will be essential towards popularizing our genetic tools and providing guidelines for their use.

