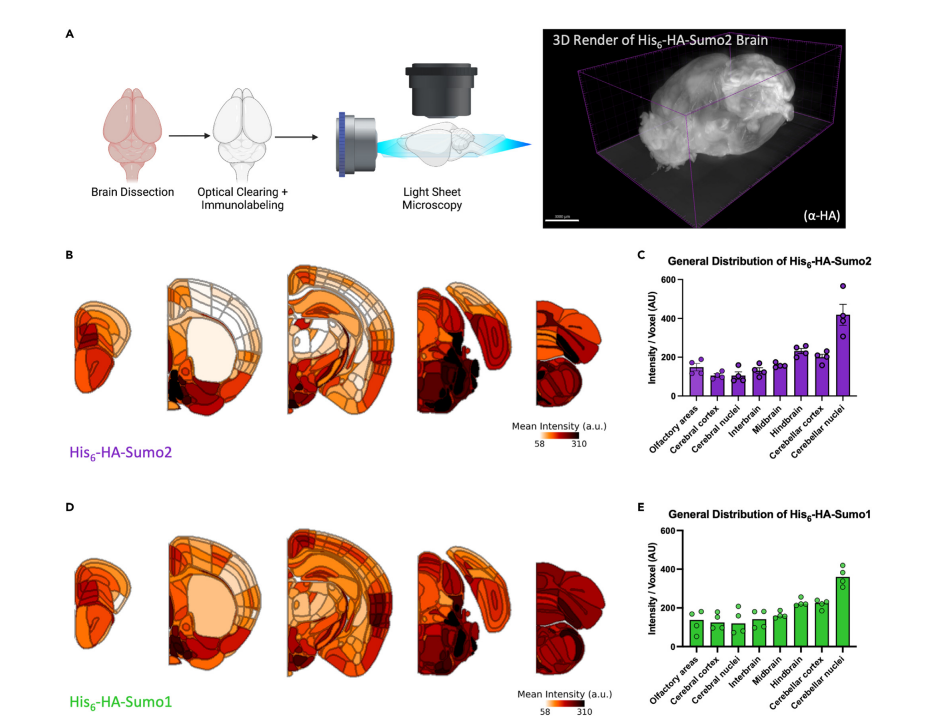
Figure 2 (Ref 1): Whole-brain imaging reveals the topographical distribution of Sumo paralogs. (A) Schematic of brain clearing and light sheet microscopy for whole brain imaging. Heatmaps and bar plots of Sumo2 (B,C) and Sumo1 (D,E) signal intensity across brain regions. Article distributed under the terms of the Creative Commons CC-BY license.
Video S1 (Ref 1): Imaris 3D render of anti-HA immunosignal in whole His6-HA-Sumo1 brain, related to Figure 2. Scale bar: 3000 μm. Article distributed under the terms of the Creative Commons CC-BY license.
Video S2 (Ref 1): Imaris 3D render of anti-HA immunosignal in whole His6-HA-Sumo2 brain, related to Figure 2. Article distributed under the terms of the Creative Commons CC-BY license.
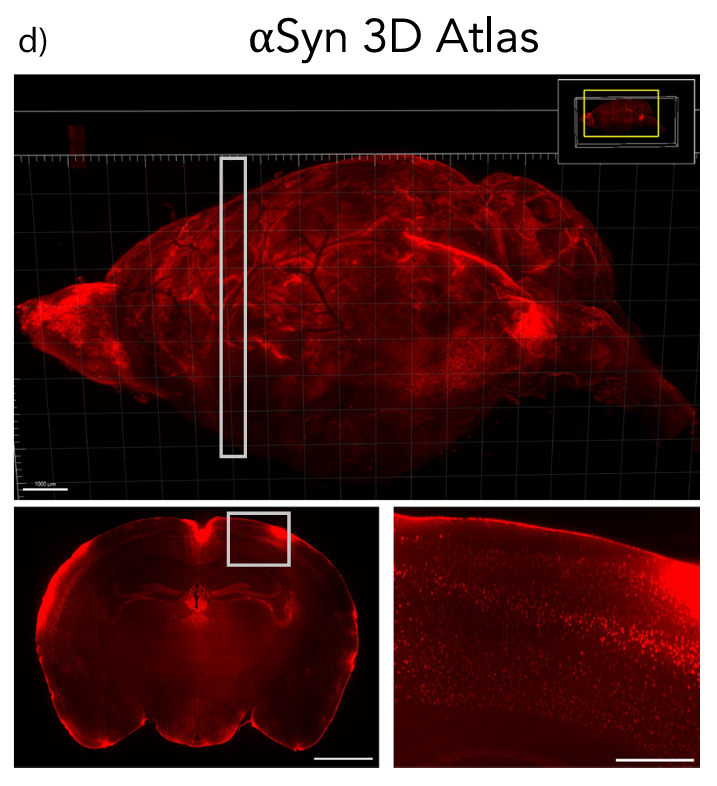
Figure 1d (Ref 2): Representative images of whole brain Flag epitope staining and imaging provide an encompassing view of αSyn topography. White boxes indicate insets. Scale bars: 1000 μm (d top, d bottom left), 250 μm (d bottom right). Article distributed under the terms of the Creative Commons CC-BY license.
For additional publications supporting this use case refer to: