LifeCanvas (LC): What areas of neuroscience are you most interested in?
Jose Maldonado (JM): I find that the scientific questions that I was interested in all revolve around circuit neuroscience. There’s a lot to learn about the molecular mechanisms that underlie neuronal processes and function, but in my view the way to increase our insight into functional neuroscience is to get a better understanding of the cellular architecture.
LC: What led you to your recent areas of research?
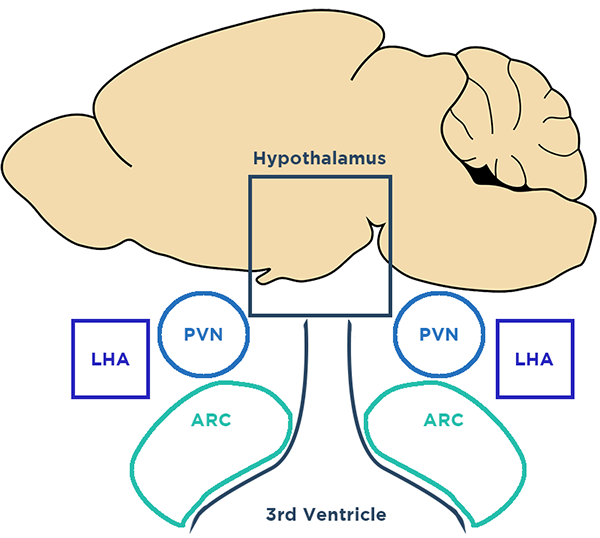
JM: When I joined Richard Simerly’s lab, he was in a field that I had no experience in: feeding. When he and I met, I realized that he was working in the kind of “arcane” areas of the brain I was interested in. I was an olfactory bulb guy after all and he had done important work in the hypothalamus. To me, the hypothalamus was almost like, “Here be dragons.”
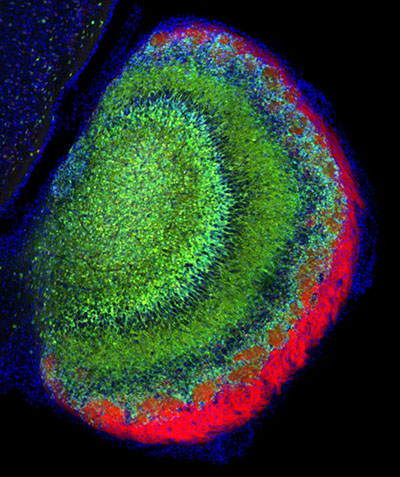
A function of that perception to me was the type of microscopy that I used when training. If you were to look at a cross-section of the olfactory bulb, you would see this beautiful structural anatomy.
I focused on the granular cell layer, which looks structurally recognizable. When you look at a cross-section of the hypothalamus with conventional microscopy, it’s a mess. Looking at that neuroanatomy in traditional cross-sectional light microscopy is like looking at anatomy through a straw. It really challenges you perceptually to imagine the cellular organization of the hypothalamus.
The ability to apply new light sheet microscopy technology to a region that was not organized in the way that I was conventionally used to appealed to me. What we weren’t expecting was the clarity, the resolution, and the speed. When we were looking at pictures of the hypothalamus from the LifeCanvas light sheet, I was trying to keep my poker face on, when what I was feeling was, “That’s what that looks like?” In the olfactory bulb granular cell layer, it’s not difficult to piece together what a 3D structure looks like from a 2D cross section. But the structural organization of the hypothalamus is difficult even for very experienced neuroanatomists like Richard to imagine based on the expression of proteins.
LC: What limitations have you encountered in your research and what tools do you think have been promising in overcoming these kinds of limitations?
JM: As a neuroscientist, I’ve had to pivot in becoming an information neuroscientist. When I got my doctoral degree, you could hide and not learn any code, and that wasn’t looked down upon. When I got back to academia, I immediately felt like a dinosaur. If you really want to address questions in neuroscience that are dependent on being able to look at the mesoscale cellular organization of neurons, you have to become relatively facile with computers and information technology.
When a group or institution gets its hands on a tool like the LifeCanvas microscope, they’re not prepared for the torrent of data. With that microscope, you have to acquire a large amount of computing power to move information. If you’re not ready to deal with fine-tuning information transport or bandwidth I/O, you need help from somebody who does, because that will be the part that makes you throw your hands in the air.
People are going to run into this IT roadblock and I’m afraid that they are going to say, “Oh, this whole thing is unfeasible.” It’s not unfeasible, you just have to get out of your comfort zone and learn (a little) Python.
The exciting thing I’m trying to push in my lab is getting out of this conception that the way to look at things is on a monitor with a little 3D box. I’m trying to look at other ways to help people visualize extremely large datasets. Big things have small beginnings. There are tools already available. But the ability to have meaningful integration requires bridging the communication gap. Neuroscientists are going to have to get better at communicating what they want to information scientists.
LC: How have the LifeCanvas team’s imaging and analysis experts been helpful as a resource and as co-collaborators?
JM: You can’t go it alone. What LifeCanvas brought was the ability to understand the problem conceptually from a software, quantitative point of view, and to be cogent partners with someone who knows what he’s getting on a granular level. The Analytics team at LifeCanvas was fantastic in getting us through the initial phase of providing the software that is now known as SmartAnalytics. The imaging team liaised between our lab and LifeCanvas so we could communicate our scientific goals.
In my first attempt to try to get this to work with some publications and Github downloads, I spent longer than LifeCanvas did, with no results. Universities have a lot of resources, but there’s not always the ability to communicate interests, and I think that’s what we got when we first started talking with LifeCanvas. In our Nature Communications paper, we adapted code and command lines that LifeCanvas generated to fit our research structure, and that very quickly resulted in a powerful demonstration of quantitative neuroscience. I looked at my publications from graduate school and thought, this would never get published today. Not only that, but I could probably knock this out in an afternoon. Why? Because we now have this very fine-tuned tool for quantitative neuroscience. That is a direct result of this collaboration between our lab and LifeCanvas.
LC: Could you discuss your work with the Vanderbilt Neurovisualization Lab (VNL) and how you act as a liaison between light sheet microscopy as a technology and the university?
You have to cut through the clutter. More importantly, it can help to better contextualize the study. My overall goal is to bridge this communication gap of how to apply light sheet to research questions in a way that helps Vanderbilt increase grant funding.
When I saw the kind of integration that we could do with the LifeCanvas products, it was pretty clear that if we could bridge the barriers for people to use this technology, we could become a more successful research community.
Light sheet allows the grant writer to convey a powerful conception in the short time you have to get something in front of a committee.
What does it take to remove all of those steps? What I’ve tried to do with the VNL is to integrate that into an operational approach. I have to help the investigator understand what they’re seeing so that they can make cogent inferences. Until it becomes easier to field and optimize at minimum a 10 GB network, people are going to need a resource like the VNL which has already oiled all the gears. People are busy being neuroscientists, and that does not involve learning how to manage large amounts of data. I think my goal with the VNL is to keep that from becoming a barrier to publishing papers or getting grants that address novel questions in systems neuroscience, until neuroscientists are ready to do so themselves.
LC: What are some of the most exciting things you've learned in general during your career?
JM: A friend and colleague’s lab managed to show that you can send information from a primate’s brain into a computer. A macaque with an implant in its brain would play a computer game with a joystick, and once the computer registered enough data about brain voltage and joystick movements, the monkey could continue to play the game without the joystick. More recently, an implant in a similar experiment was adjusted to a sensory motor. When a digital hand went over a certain pattern on the screen, a signal was sent into the chip in the monkey’s brain that triggered a “rough” sensation. The monkey was able to differentiate between sensation types within the touch sensory paradigm without actually sensing anything directly. What does this mean for what we might eventually call a natural extension of ourselves?