What is a brain atlas?
A brain atlas is an image of a brain with all of the regions individually labeled. Atlases are species-dependent: there are mouse atlases, rat atlases, human atlases, and more. Brain atlases can either be 2-dimensional (made of annotated brain slices) or 3-dimensional (comprising a 3-d image and 3-d annotation of a full brain.)
What is the Allen Brain Atlas?
The Allen Brain Atlas, also referred to as the Allen Mouse Brain Common Coordinate Framework version 3 (ccf3 or ccfv3), is a specific mouse brain atlas created by the Allen Institute. It is the most widely-used atlas for the mouse brain. The most recent version, ccf3, was created by averaging brain images from slices acquired from 1,675 mice. It delineates 43 isocortical regions, 329 subcortical regions, 8 ventricles, and 81 fiber tracts, and is open access and easily accessible.
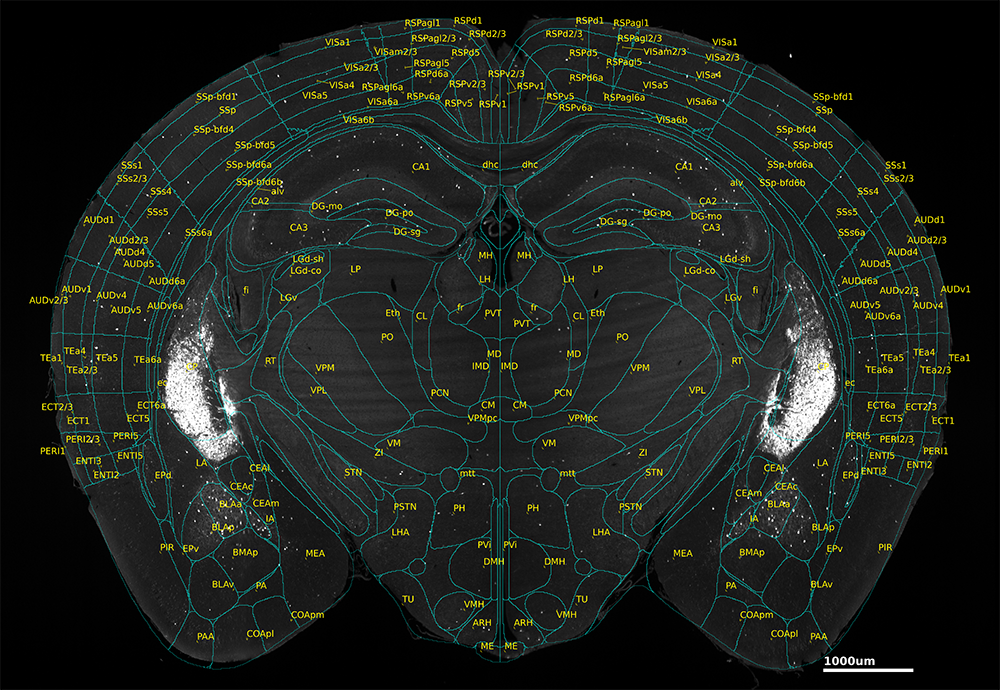
While it was created from 2-d slices, these slices were combined to provide a full atlas of the entire 3-d structure of the mouse brain. This makes the Allen Brain Atlas an incredibly powerful tool for neuroscience research, enabling researchers to discuss their findings in a common coordinate system.
What is atlas registration and why is it important?
Atlas registration is the process of matching the image of a particular experimental sample to a brain atlas. Because no two brains are exactly the same, images typically can’t simply be superimposed onto the brain atlas. Instead, either the image of the sample or the image of the atlas must be slightly warped in order to get good alignment – for example, slightly stretching or expanding one or another region.
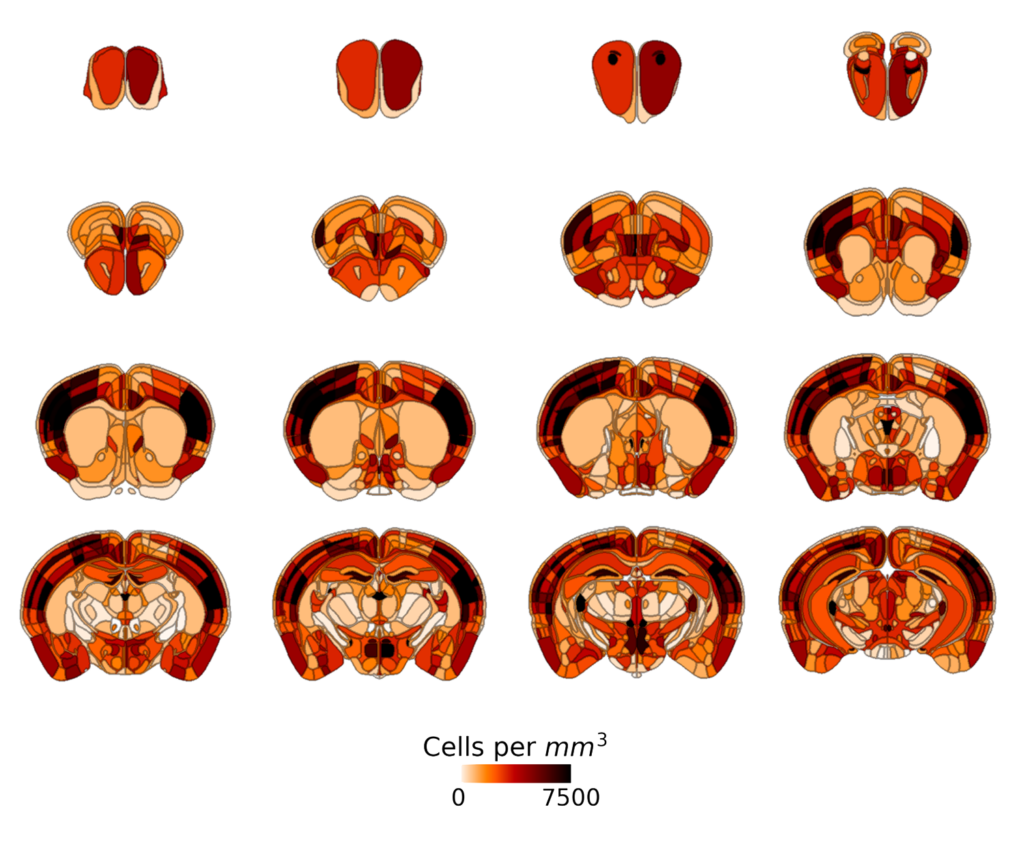
By finding a way to warp the images such that your sample superimposes perfectly onto the brain atlas, you can then use information from the atlas to identify brain regions in your image. This can make it simpler to make sense of your data by examining effects by brain region and comparing effects between regions. Without registration, it becomes difficult to impossible to get fine-grained information about region-to-region variations in your data.
How do you register brains to a brain atlas?
The most common method of registering brains to an atlas is to solve an equation which tells you how to warp coordinates from the brain into coordinates on the atlas (or vice versa). This process often takes multiple steps.
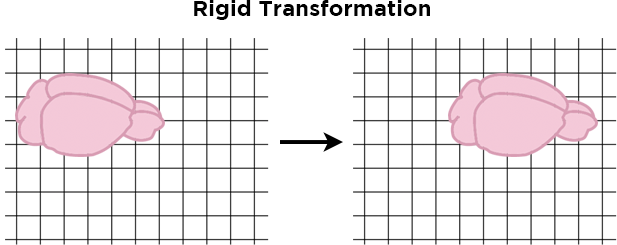
First, a rigid transformation shifts the images in the x,y, or z dimensions in order to find the best alignment, but does not change the size or shape of the images.
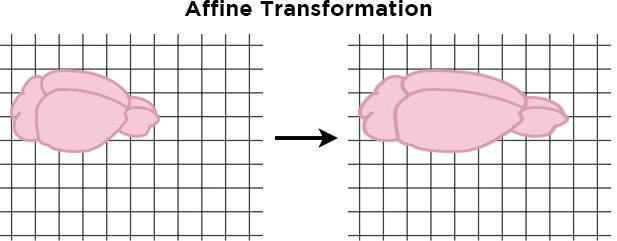
Next, an affine transformation stretches or shrinks the images uniformly along a single line – for example, it could stretch the sample front-to-back to elongate the sample, while keeping its width the same.
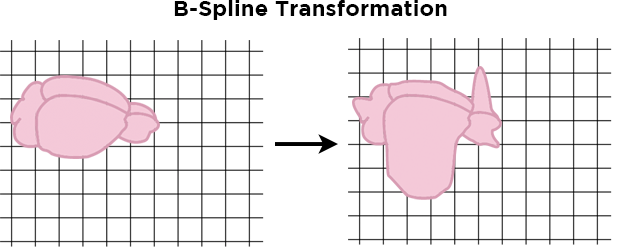
Finally, a non-linear transformation such as a b-spline transformation can stretch and shrink individual areas of the sample to further tweak the alignment.
To perform these transformations, researchers often use pre-existing code packages. Two popular code packages for brain atlas registration are Elastix and the Advanced Normalization Toolkit (ANTs), both of which interface with the Insight Toolkit (ITK) in order to perform registration between images, such as between a brain atlas and a sample image, or between two different experimental images. There have also been recent developments from researchers trying to leverage novel deep learning approaches for atlas registration, such as BIRDS and MesoNet.
Atlas registration can be done in two dimensions by matching a single brain slice image to a single brain atlas slice, or in three dimensions by matching an entire brain or hemisphere image to a 3-d atlas. Computationally, two-dimensional registration is somewhat simpler, but it is unable to account for issues that could arise from the slice coming from a slightly different part of the brain than the atlas, because it does not have access to information about the third dimension.
What are common registration issues and how can you address them?
While simple in concept, atlas registration can be tricky to master. If at the end of atlas registration, you determine that the atlas and sample are imperfectly aligned, you have a few options. First, you can consider the parameters of the registration algorithm. Sometimes you can achieve better results by testing different parameters.
"Without registration, it becomes difficult to impossible to get fine-grained information about region-to-region variations in your data."
Second, you can add in a manual registration step, where you manually inspect the registration and make modifications to improve it. This can be done through graphical tools designed for the task, such as the NUGGT (NeUroGlancer Ground-Truth) program.
Finally, you can consider how well your sample correlates with your reference atlas. Because most if not all registration algorithms examine the correspondence between signals for the atlas and sample as their point of reference, it helps to have your image contain data that is as similar to the atlas as possible – for example, if you are using the Allen Brain Atlas, their images show autofluorescence signal, so the algorithm will perform best if you use an autofluorescence channel from your sample for registration.
If atlas registration still seems daunting, LifeCanvas can help!
LifeCanvas’ SmartAnalytics software program performs automatic registration for you and interfaces with nuggt to enable manual verification of registration results, allowing you to perform atlas registration in-house with no coding required. If you’d rather let us take care of every step of the process, you can send samples to us, and we will clear, label, image, and analyze them (including atlas registration) through our CRO services. Contact us to speak to one of our experts about how we can help you supercharge your experiments!

