Neuroscientists studying major neurological disorders such as Alzheimer’s, Parkinson’s, and Huntington’s diseases frequently use transgenic mouse models to evaluate disease progression and the impact of potential therapies. For instance, mice with overexpression of Alzheimer’s related proteins (e.g. amyloid precursor protein, APP) or mutations that trigger pathology (see Jankowsky and Zheng 2017) demonstrate the hallmarks of disease both behaviorally and neuroanatomically.
In collaboration with Taconic Biosciences, LifeCanvas has embarked on a thorough characterization of Alzheimer’s disease development in two key mouse models: Taconic’s ARTE10 #16347 (APP-PS1) and APPSWE #1349 lines.
Effective assessment of diseased tissues in these models requires both high resolution visualization of neuronal damage markers like plaques and neurofibrillary tangles, and accurate localization and quantification of neuroinflammation responses, such as clustering of reactive astrocytes and microglia.
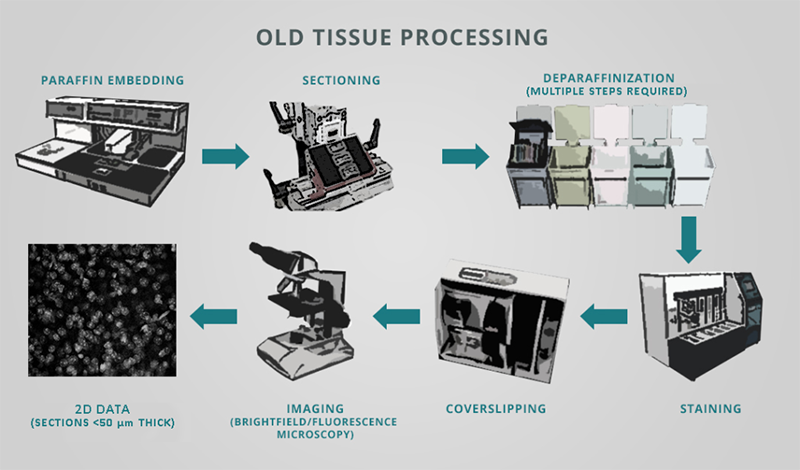
Researchers commonly use classical immunohistochemistry (IHC) to locate and quantify aberrant protein aggregates and cellular responses to neurological disorders in brain tissue. However, the traditional tissue histology workflow is not only error-prone and expensive, but also slow, inefficient, and laborious. IHC generally calls for embedding fixed brain samples in paraffin, painstakingly sectioning and mounting sections on slides, deparaffinization, staining with antibodies or dyes, and imaging all of the sections.
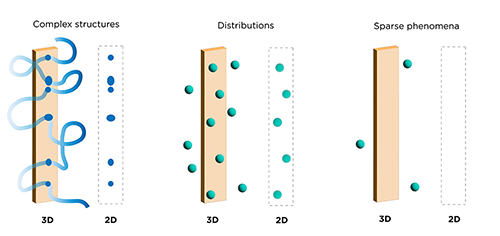
Computer reconstruction of 2D image stacks has enabled 3D rendering of these datasets, however critical information is often lost. Physical slices can be damaged during tissue slicing, preparation or processing. Even if a slice makes it to the microscope, often only a subset of slices are selected for visualization and analyses, as it is prohibitively time-consuming to image every section. This can introduce several problems such as: inconsistencies between sections, overlooking rare populations of cells, disrupting and misinterpreting continuous structures like vasculature and neuronal connectivity, and inaccurately quantifying complex spatial relationships between specific cell types (see Liu et al. 2021).
In researching neurological disorders, spatial information is key to assessing the interactions of reactive glia (e.g. astrocytes and microglia) with amyloid plaque depositions.
The extent of infiltration and clustering of reactive glia into plaque regions can reduce amyloid load and thus potentially ameliorate symptoms – a critical endpoint for a therapeutic study (Wang et al. 2020). For instance, the surrounding of amyloid plaques by microglia has been reported as an important mechanism for minimizing plaque toxicity and reducing neuritic dystrophy (Condello et al. 2015). Microglia may also play a critical role in converting diffuse amyloid plaques to more compact plaques that are less neurotoxic and block disease progression (Casali et al. 2020).
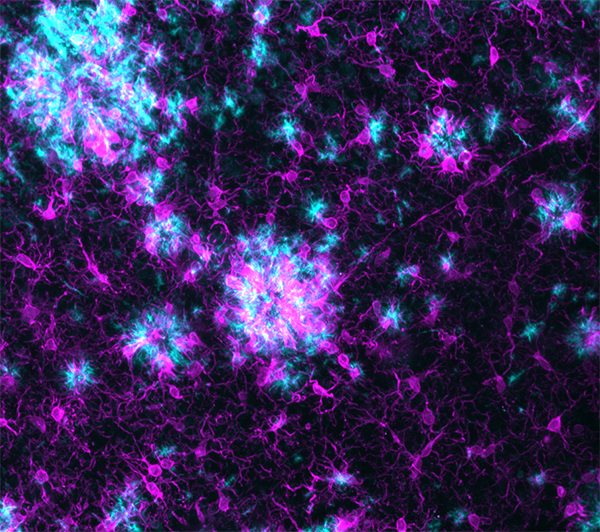
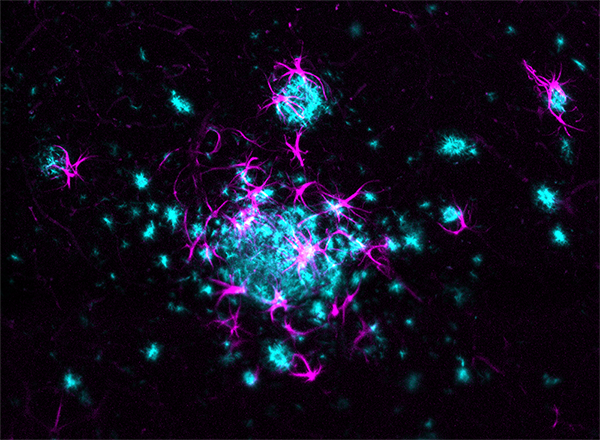
LifeCanvas’ collaboration with Taconic Biosciences has produced compelling images and analyses of disease progression and neurological response in Alzheimer’s mouse models. 3D renders of images from our lightsheet microscope SmartSPIM reveal reactive glia (astrocytes and microglia) clustering around and infiltrating amyloid plaques in exquisite detail.
To obtain and analyze these data, whole brain samples from model mice are processed through our complete pipeline:
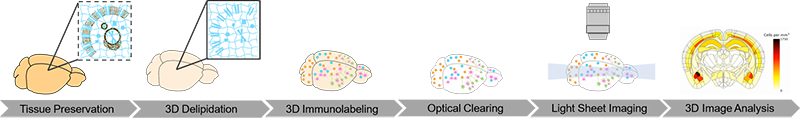
- SHIELD preservation
- Clear+ delipidation via SmartBatch+
- Immunolabeling with antibodies against beta-amyloid (to stain plaques) and glial markers IBA-1 (for microglia) and GFAP (for astrocytes) via SmartBatch+
- Refractive index-matching with EasyIndex
- Light sheet imaging with SmartSPIM microscope using 3.6X objective for whole brain and 15X objective for higher resolution imaging of select ROIs
- Image registration to the Allen Brain Atlas with regional cell and plaque detection via SmartAnalytics
As plaque morphologies are critical for understanding disease progression, neuroscientists need accurate, information-rich image datasets to evaluate the 3D environment of plaques and glia within affected brains.
Employing Taconic Biosciences’ robust and reproducible murine models for Alzheimer’s disease and LifeCanvas’ 3D histology platforms, researchers can explore critical details of plaque structure and interactions of neuroinflammatory responses.
By better understanding disease progression and exploring potential therapies, our partnership has considerable promise for enabling investigators to more thoroughly address neurological disorders.

