Breathtaking progress in life sciences has brought us innovations such as high-throughput and individually-affordable genomic sequencing as well as next-generation flow cytometry that can phenotype dissociated cells for their expression of dozens of markers simultaneously. The development of cutting-edge technology solutions with standardized workflows has led to increased efficiency and reliability for these and other cell-based techniques. Understanding how populations of cells in the native tissues they form are altered in disease states is an equally important venture and unfortunately is one that has seen relatively little in the way of breakthrough technology development in recent decades.
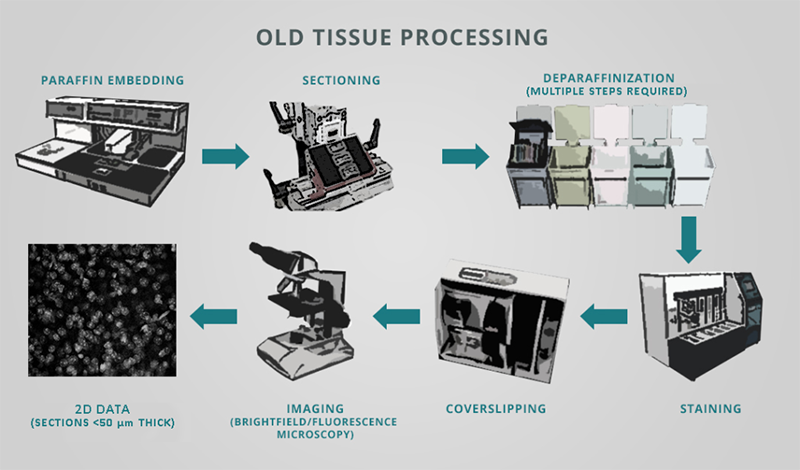
Specifically, the classical histology workflow centered around the equipment shown in Figure 1 is still in widespread use today in academic labs, biopharma companies, and healthcare centers. This process involves:
- Cutting tissue into thin sections, typically ~35-40 µm thick (but down to ~3 µm which can require paraffin embedding)
- Mounting the sections onto glass slides (and performing subsequent deparaffinization)
- Labeling these sections with classical stains or antibodies
- Visualizing the sections either in their entirety at low magnification or by investigating discrete regions of interest (ROIs) at higher magnification
More recent advances in optics and camera technology have allowed researchers to store microscopic images for offline analysis, and similar developments in fluorescence detection methods have facilitated automated analysis of signals of interest. Thanks to confocal microscopy and 2-photon excitation it has also become possible to extract some limited 3D information from thin slices and somewhat larger tissues (up to ~0.5-1.0 mm thick), although labeling is precluded in the latter especially when using higher molecular weight reagents such as antibodies.
However, the early part of the histology workflow, before the images are acquired, has remained fundamentally the same for some time, with tissue still needing to be thinly sectioned for it to be labeled and visualized. While multiple samples can be sectioned at once and then batches of slides can be stained in parallel by incorporating a bit of automation, this process has remained:
Laborious, because human control is required for sectioning & mounting of the tissue
Error-prone, because sections can be damaged or lost during handling
Inefficient & expensive, because large quantities of labeling reagents can be needed to batch-process large numbers of slides, like are generated from sectioning an organ such as a mouse brain from end to end
Slow, as labeled sections are imaged individually and then handled separately
Further, in this traditional workflow, classical stains and especially immunohistochemistry only label the most superficial regions of the tissue. This reality, along with the limited penetration depth of light into tissue as it has typically been prepared, are together the main reasons why tissue sectioning is performed. Additionally, because it is prohibitively time-consuming to image the entirety of a set of sections comprising a given sample/organ, the results produced from this workflow often rely on sampling principles that can introduce inaccuracies or cause intra-area differences that are biologically meaningful to be missed.
Tissue Clearing, a 21st Century Approach to Histology
In the past 10 years, however, and especially since introduction of the CLARITY technique in 2013 (Chung et al., Nature), a revolution has occurred in the field of tissue processing that has catapulted the discipline into the 21st century. Today, thanks to the chemical engineering-inspired approaches of CLARITY and more recently of SHIELD (Park et al., Nature Biotech, 2018), thick tissue samples the size of intact rodent organs and beyond no longer need to be meticulously cut into hundreds of sections to enable imaging and even labeling to take place.
By preserving samples to withstand removal of all lipid-filled cell membranes ‒ which scatter light ‒ from the tissue, the samples can be rendered translucent in a process called delipidation that involves detergents and electrophoresis. Following delipidation of a sample and its subsequent incubation in a solution that raises and homogenizes refractive index throughout the tissue, the sample becomes optically transparent and can be imaged in its entirety while completely intact and un-sectioned. To learn more about these techniques, please visit our Technology page.
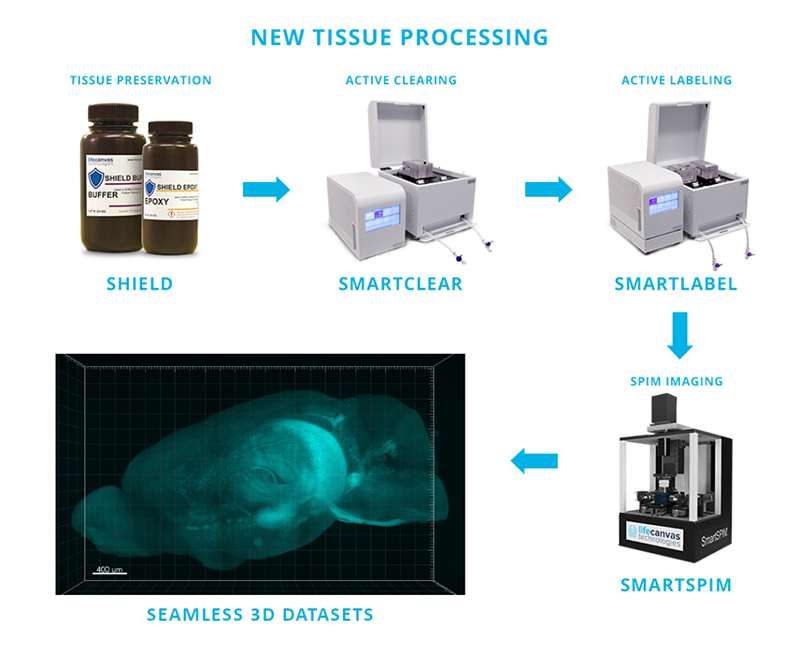
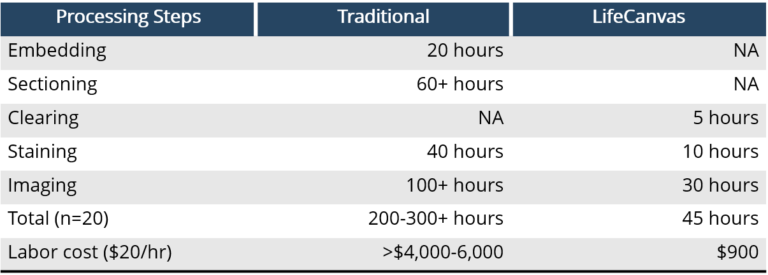
In addition to offering researchers a new and valuable perspective on histology, this 21st century tissue processing workflow requires dramatically less hands-on time vs. traditional sectioning-based approaches. Using the products below in LifeCanvas’s end-to-end, sample-to-dataset pipeline (Figure 2), entire rodent organs can be:
Actively labeled with antibodies & other molecular probes
Light-sheet imaged at single-cell resolution
All with minimal user intervention and with labor requirements that are a fraction of traditional histology workflows.
With LifeCanvas’s pipeline you can not only simplify your workflow, but also take advantage of the power of whole-sample methods to:
Reduce processing work and view samples in multiple anatomical planes
Localize regions of interest more confidently by visualizing organ-sized datasets that provide greater context
Acquire data on all sample regions in parallel, facilitating analyses that are more quantitatively robust
Explore fertile ground where novel and unexpected discoveries can take place
Learn more about how whole-sample analysis techniques can facilitate and deepen your research by reading our earlier blog post.
Interested in using LifeCanvas’s CRO services to expedite your research? Our lab can preserve, clear, label, and image intact samples with little turnaround time! We also offer analysis solutions including cell counting and atlas registration.